Happy Birthday, EaSeq
Happy Birthday, EaSeq
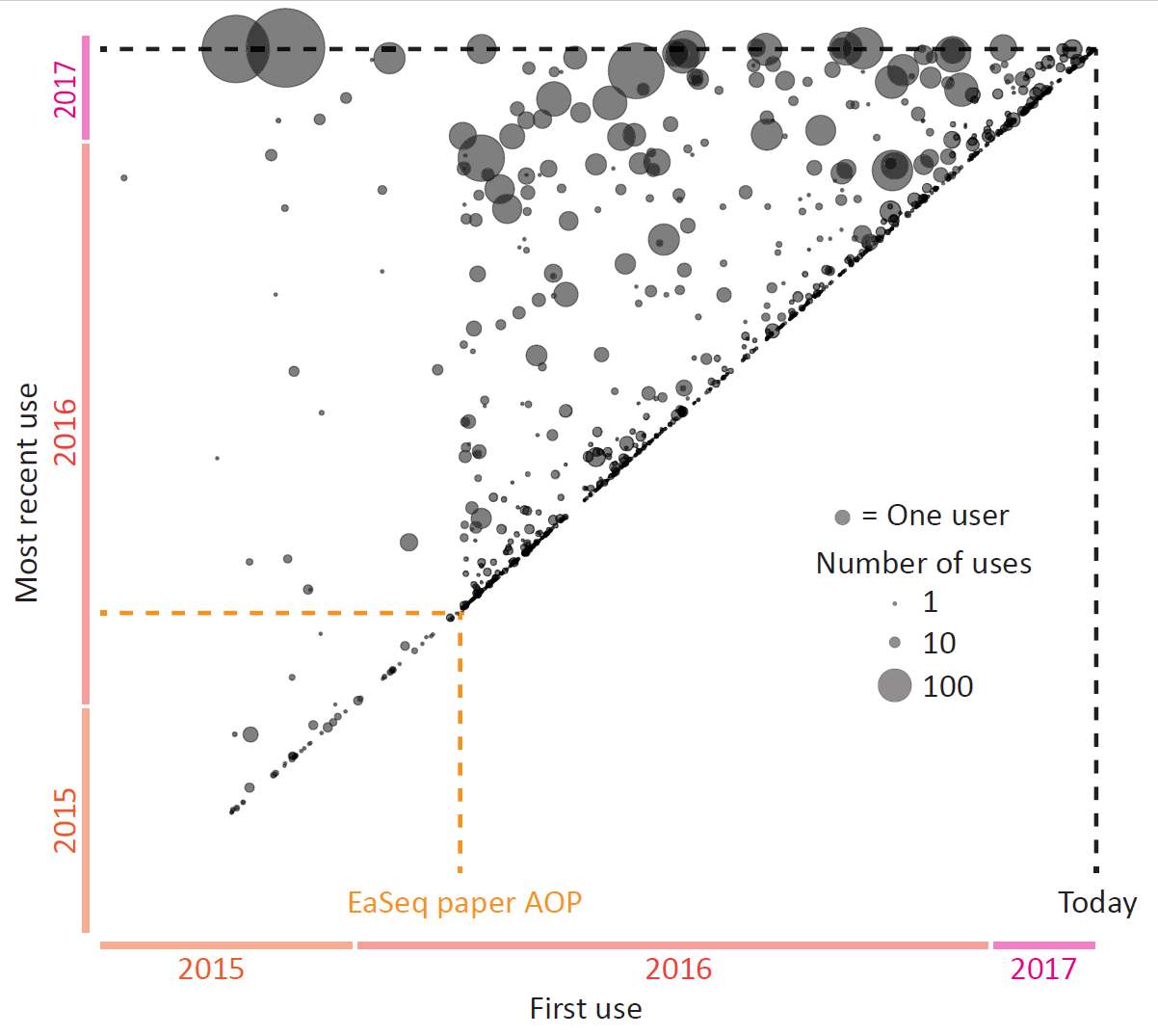
Today EaSeq turned one year old, counting from the day our paper went AOP.
I have looked a bit into the statistics to get an impression of how many people (or more correctly: computers) that have been using EaSeq during the last year.
There is a lot that I cannot infer from the available data. But it is clear, that apart from me, then there is a substantial number of people who have been installing EaSeq (>2,500 installations on >1,000 different PCs), a steady influx of new users, and quite a lot who have been using it persistently (Data are cleaned for my own use btw.). One of the big circles in the upper left corner are the guys in the Helin lab and the other is a French guy in Canada who have managed to use EaSeq in ways I never anticipated. I hope that some of you newer users will soon be able to surpas them 😀
Honestly, this feels very fulfilling and heart-warming. Thank you very much for the faith that you have shown. It is one thing to publish science, but it is even more satisfactory to know that the work does make a difference for other people – which is probably also why many of you chose to study diseases…
Unsurprisingly, there is a large fraction users who install EaSeq and then abandons it. This can happen for number of reasons: (the good) the user migrates to a more powerful PC, (the bad) it does not really cover the areas that are needed or expected, and (the ugly) the user decides that it is too complex to use or too hard to learn using the program.
It is probably common for most freeware programs that people try it out and move on. However, if you install EaSeq but decides against using it, then I would love to learn why – it would really be ignorant of me not to listen to your reasons and try to improve EaSeq accordingly. Therefore, please do let me know why if you decide not to use EaSeq after installing it.
To all of you – regardless of whether you hang on or not – thank you for trying EaSeq, and good luck with your projects!
Congratulations to our users! v1.04: PDF export and other plot improvements