THE LATEST NEWS
Workshops

New position
Upcoming maintenance
Congratulations to our users!

What an excellent week for some of our users (and thereby for EaSeq too): Three new stories using EaSeq came out.
Congratulations to these users and their colleagues!
In case that some of you seek inspiration on how to use EaSeq, then take a glance below and in the list of papers made using EaSeq:
Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization
Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG Cell. ... Read More
Happy Birthday, EaSeq
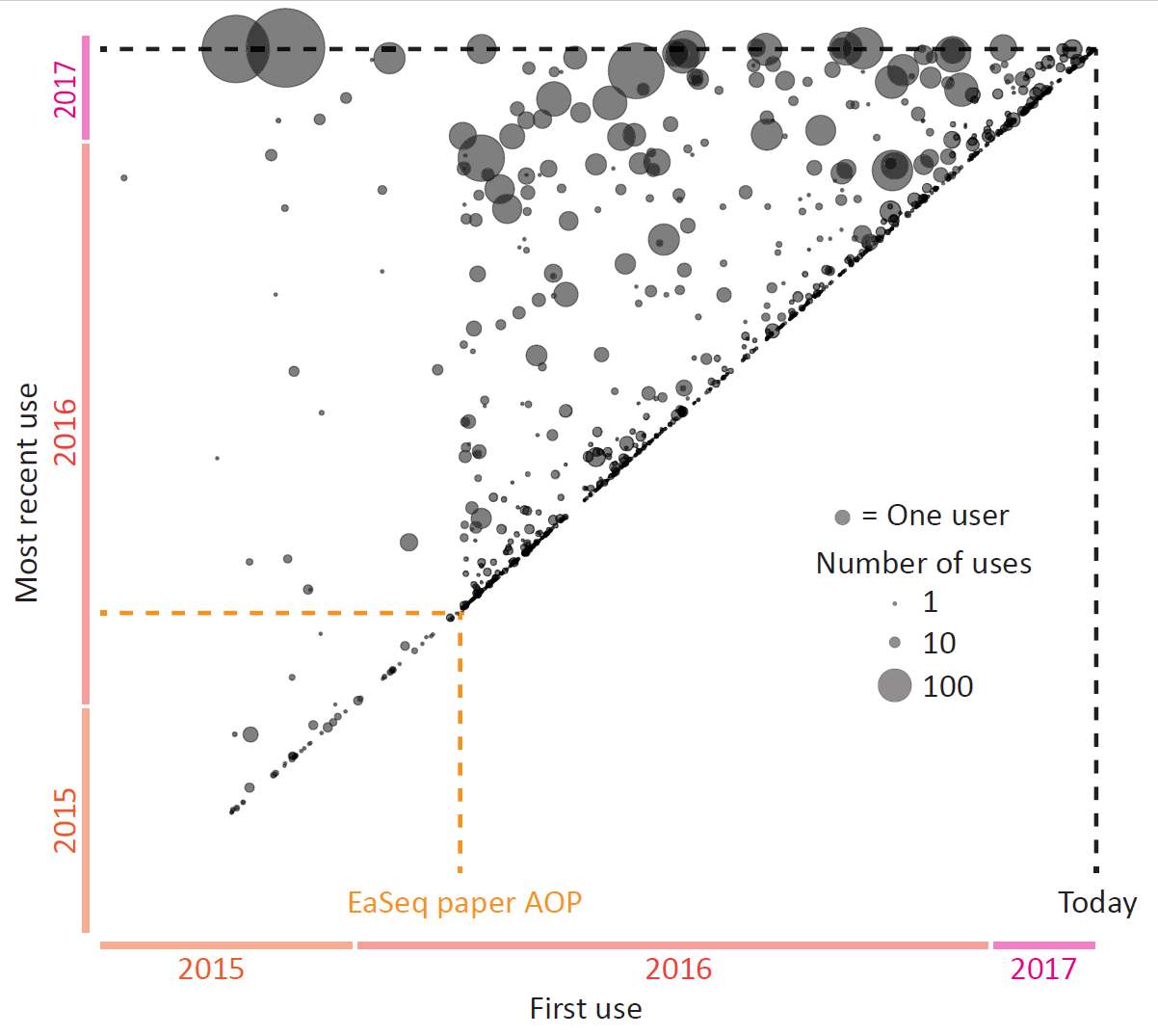
Today EaSeq turned one year old, counting from the day our paper went AOP.
I have looked a bit into the statistics to get an impression of how many people (or more correctly: computers) that have been using EaSeq during the last year.
There is a lot that I cannot infer from the available data. But it is clear, that apart from me, then there is a substantial number of people who have been installing EaSeq (>2,500 installations on >1,000 different PCs), a ... Read More
v1.04: PDF export and other plot improvements
v. 1.04 is out, and while it might not be obvious at first glance, much was changed under EaSeq’s hood when going from version 1.03 to 1.04.
The three most important improvements are:
1) Export plots as pdfs.
I had to make a substantial redesign of the algorithms underlying the plotting part to allow support for pdf output.
The new version has been throughoutly tested and debugged for a couple of months, and to my knowledge, the plots perform ... Read More
New features for beta testing

The most recent version of EaSeq (v. 1.03) got a new ‘Beta-panel’ (download here).
It can be found in the right side of the Labdesk, below the button for the Navigation panel, and so far it contains a couple of new tools:
The ‘Get Sequences’-tool.
This tool downloads genome sequences (requires some GBs of disk space) from UCSC and export the sequences from a regionset. This is useful if you would like to generate input data for motif finding at a set of peaks ... Read More
ChIP-seq data analysis workshop on April 12
Would you like to learn how to analyze, explore, and visualize your own and published genome-wide ChIP-seq data?
Then you might be interested in the upcoming workshop ‘ChIP-seq data analysis and visualization for life scientists’, introducing the software environment EaSeq (https://easeq.net/).
EaSeq was recently published and is free of costs, runs on a typical PC, and enables analysis through a graphical user interface. So no need for programming skills or expensive equipment – only basic biological and genomic understanding.
It combines the interactive locus-based exploration ... Read More
New tutorial videos
Sessions files for our paper are available now
EaSeq manuscript published
Wanted
Running EaSeq on a Mac or Linux machine

Poster

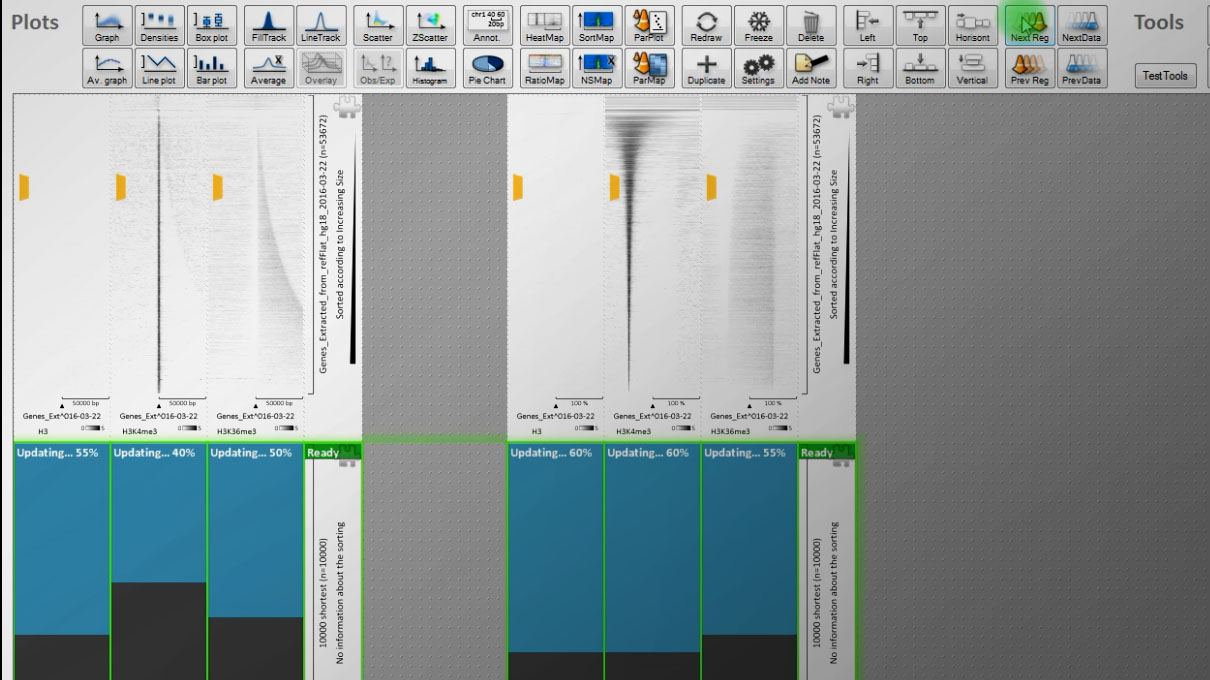
A paper worth of figures in 30 minutes?
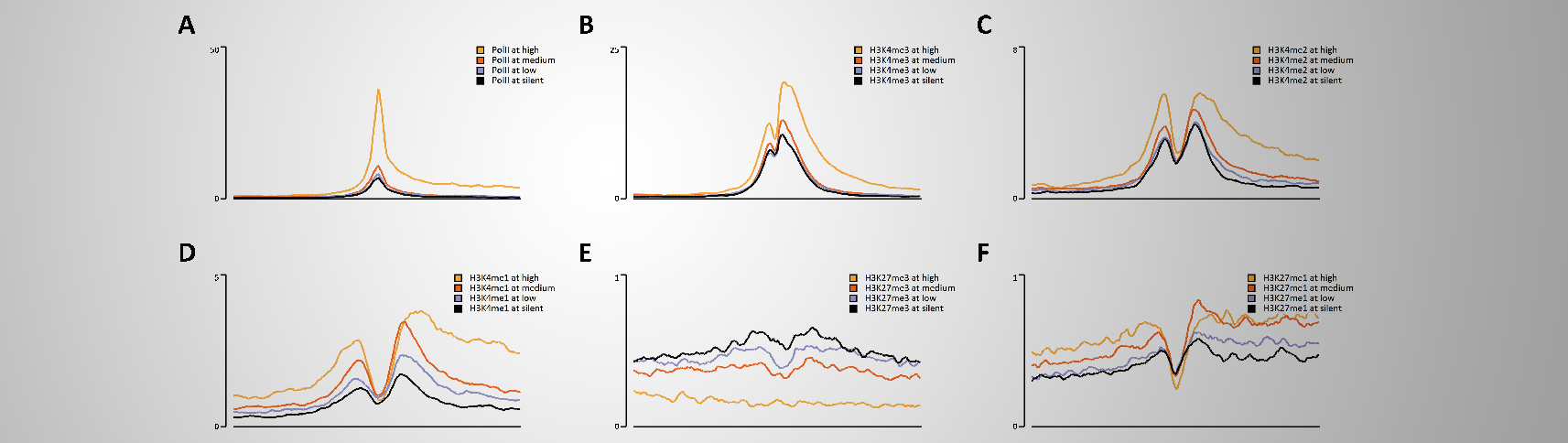
It might be bold to state that EaSeq can accellerate the process of turning comprehensive ChIP-seq datasets into interpretations and figures.
We therefore put this statement to test by remaking the 34 figures and ChIP-seq analyses from Barski et al. (Cell, 129(4):823-37).
Altogether, this took half an hour(*) just by pointing and clicking (see output here), and we believe that this pace is indeed a welcome improvement in analysis time. Especially considering that no programming was involved.
(*) Admitted: We did pause between each figure, planned ahead, and the ... Read More
v. 0.97 is out
We are happy to release v. 0.97. The user experience has been improved markedly, so we recommend upgrading older versions of EaSeq to v. 0.97.
Among other things, the GUI controlling the plot handling has been completely remade from scratch, and now runs more efficiently and smoothly. It has been tested in a range of conditions for a while, but there might still be a bug or two hiding. So please let us know if it misbehaves in your hands.
The Tutorials were affected by this ... Read More
What is cooking?